Service
Comprehensive Virus Screening
Technology
RNA Sequencing
Target Crop
All
About the test
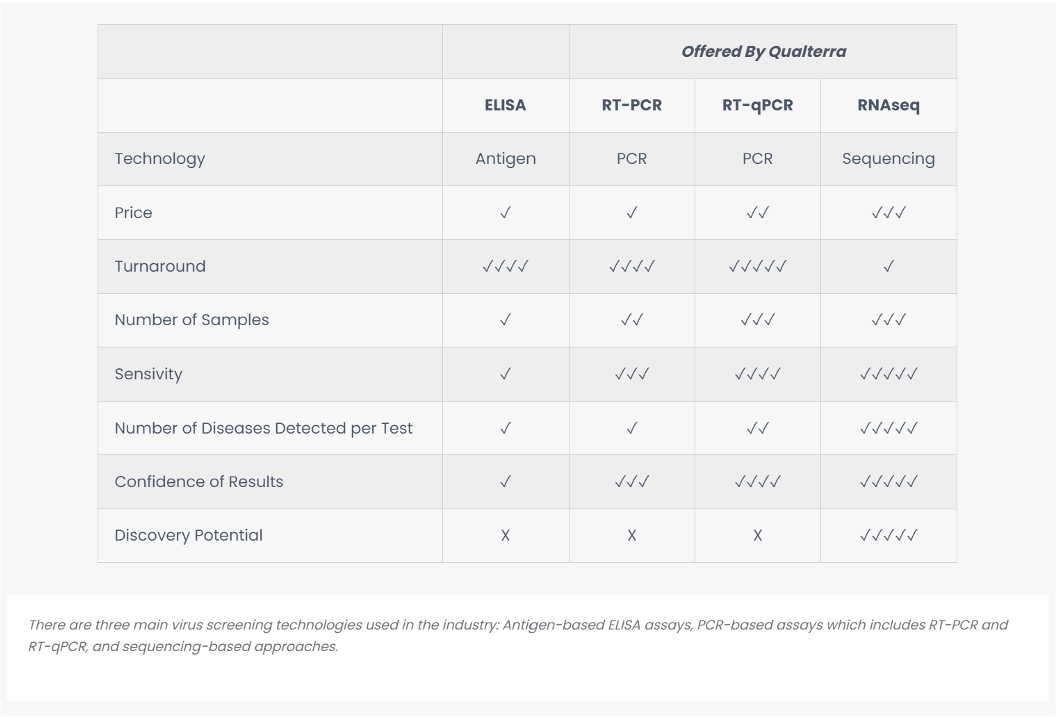
Our most sensitive option for detection of viruses and viroids, Comprehensive Virus Screening combines state of the art viral enrichment with next-generation RNA sequencing technology. Virtually all viruses and viroids present in the sample are detected, even at very low titers and before infected plants become symptomatic. This method is ideal for screening mother plants, new varieties, and pooled blocks of crops. Our technology is crop-agnostic: we have extensive experience in horticultural crops, but any plant can be tested for viruses using this method.
A collaborative approach. We are working together with partners in the industry to bring cutting edge science to the horticultural sector. As part of Qualterra’s continuing mission to innovate, Qualterra has been awarded Small Business and Innovation grants to compare several Little Cherry Disease (LCD) detection methods and develop high throughput methods for sensitive, quick turnaround and cost-effective detection.
Comprehensive Virus Screening starts with RNA from the plant tissue that is being screened, which contains both the plant’s native RNA and RNA from any other organism that was in the plant (including pathogens like viruses, viroids, or phytoplasma). We enrich for pathogenic RNA, and then the RNA is broken into smaller pieces. The sequencer then “reads” the sequence of each piece of RNA, producing a library of sequences. We can search the sample library against a database of known plant pathogenic virus sequences; if there are any matches, the sample is infected with that virus. We line these sequences up against the known genome of the virus, to visually confirm the infection. If possible, we also confirm the infection by performing a RT-PCR based assay targeting that virus.
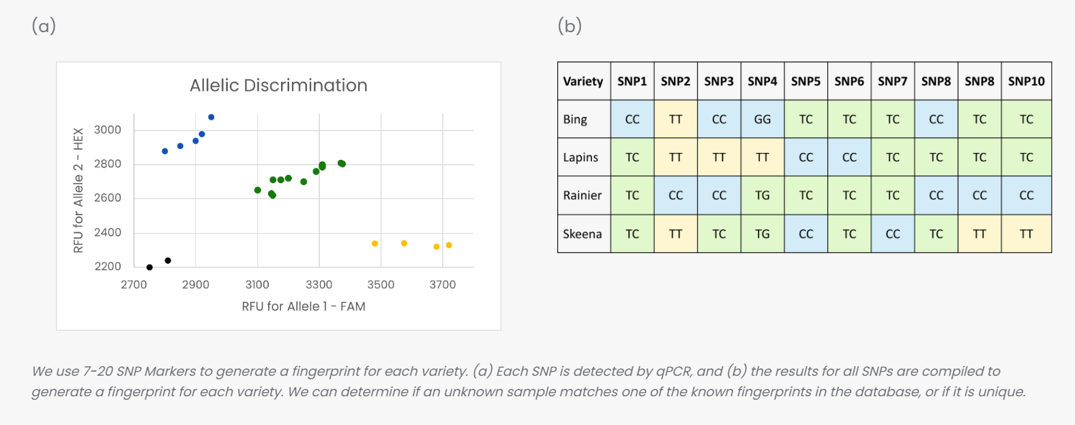
We have curated a list of over 2,500 viruses, viroids, phytoplasma, and common diseases. This database contains all plant pathogenic viruses found on the NCBI database, and we have added phytoplasma and other diseases that are commonly tested for in our area.
Check out the full list in this Excel spreadsheet: [link to download Qualtera Virus Database 2023]. (Excel, 65kb)
Hint: use Ctrl+F or Cmd+F to search for the full species name of your pathogen of interest.
If your disease of interest isn’t on the list, let us know! We’d love to hear more about your project.
Comprehensive Virus Screening tests for all 2,500+ known plant pathogenic viruses, plus other common pathogens, and is the most sensitive detection method available. It is most useful for testing mother plants, new varieties, and pooled blocks of crops, or when you believe there is a complex or unknown infection in the crop.
Targeted Virus Screen is a more economical option to test for the diseases most commonly found in the crop of interest. We offer panels curated for cherry, pome (apple and pear), or hop.
This is most useful in cases where you see symptoms in the field and have narrowed down the cause to one or a small number of viruses.
Decreasing costs through pooling: A great way to reduce the overall cost of testing many samples is to combine several plant samples together in the same test. For example, it’s common to test 5 trees together. If the pool is clean, then you know all 5 trees are virus free. If the pool is infected with a virus, you can either consider all the trees to be infected, or you can follow up with additional testing to determine which tree(s) caused the positive hit.
Possible strategies to consider:
1) Pooling is most common if you have a group of plants that you think are all clean, or if you are testing many plants and it would be cost-prohibitive to test individually. We recommend pooling 5 plants per pool, but you have as few as 2 or as many as 10 plants in the same pool. The more plants per pool, the less sensitive the test – but even with 10 plants in a pool, Comprehensive Virus Screening is still significantly more sensitive than RT-PCR.
2) Alternatively, no pooling is recommended for screening of important plants: mother plants or new varieties which will be propagated from, where it’s vital to mitigate the risk of propagating infected material.
How to pay through our online payment platform and submit pooling information: When you check out online, you are ordering the number of tests you want to run – not necessarily the number of plants/samples you want tested. If you want to test 20 plants in pools of 5 plants each, that means you want to request 4 pooled tests – so you would check out and pay for 4 tests. We will provide you with a sample submission form, where you can list all the samples and how they should be pooled.
Feel free to contact us – we are happy to help set up a sampling plan that will address the concerns you have.
We will contact you with sample preparation and shipping information after the test is ordered, but here are some pointers to get you started:
When to test: The best time to detect viruses is when they are actively replicating in the plant – and this can vary based on the virus you are targeting! A good rule of thumb is that June-August is the best time to test for most diseases, including testing for the viruses and phytoplasma that cause Little Cherry Disease.
Harvesting and Submitting Samples: Plan to use clean pruners for each separate tree/plant, and keep material from each plant separate in its own bag, even if samples are being pooled for testing. Provide 6” of stem/branch and 3-5 mature leaves for each plant. Keep all material cold, and ship as soon as possible via overnight courier.
What’s the turn-around-time?
We do all sample prep, sequencing, and analysis in house, so we can usually provide results in 2 weeks.
.


