Service
Variety Verification
Technology
Variety Verification
Target Crop
Apple
About the test
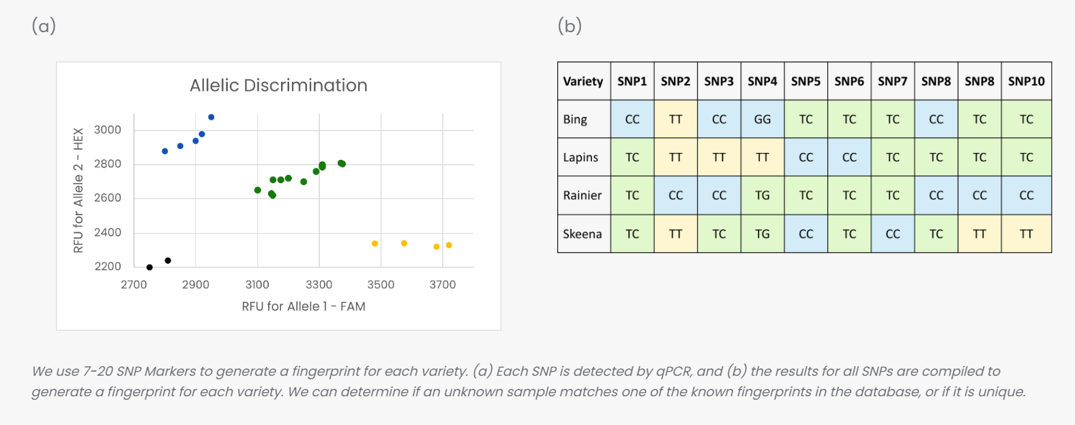
Our testing methodology uses Single Nucleotide Polymorphism (SNP) genotyping. SNPs are unique genetic markers which allow us to create a distinctive “fingerprint” for each plant variety. By comparing these fingerprints, we can accurately determine if two plants belong to the same variety.
Qualterra maintains a SNP database for Apple, Cherry, Grape, Hops, Pear and Strawberry, which are continually updated to incorporate new cultivars.
SNP Fingerprinting tests between 7 and 20 SNP markers. For each SNP marker, the plant genome contains two copies of that marker, the same way that humans have one copy of their genome from their mother and one from their father. In the case of SNP markers, there are generally two variants (“alleles”) of the marker that the plant could receive, which we could call A and B. So, for each SNP marker, a plant could have inherited an A from the mother and an A from the father giving it two A’s (“AA”), or it could receive two B’s (“BB”), or one of each (“AB”). We can detect the three options versions AA vs. BB vs. AB using qPCR for each marker, and together these results produce a fingerprint for each variety.
Mixups can come in many different flavors – and your exact situation will determine how to approach the problem. Here are some pointers:
- Each sample you send in should come from a single tree (samples cannot be pooled or mixed from different trees).
- If you believe that an entire population (e.g. a block) is the same variety, we recommend testing 3 samples from that population.
- If you have identified that some plants within a block/population are different than others, we recommend testing 3 plants from each type.
- You can add additional samples to gain insight on the situation: how large the mixed-up area is, where the border is, or to gain confidence in determining what the composition is (e.g. 5% plants are incorrect vs. 50% plants are incorrect).
- If you are trying to determine if a block/area is infected or not, we generally recommend sending in up to 30 samples from a single area; 30 allows you to find a mixup of 5% with 95% accuracy.
- Label the trees that were sampled from (e.g. numbering 1, 2, 3… works well). Use accurate GPS coordinates, or mark them on a map and also label the tree. This will help you to correlate the results back to the field and make management decisions.
- Send in a reference if needed. Keep in mind that Variety Verification using SNP fingerprinting works by comparison – so double check that the varieties you want to compare are in our reference database. If a variety you want to compare against is not listed, you will need to provide us with samples of the variety (contact us if you need assistance). See the “Do I need to include a refence to compare against” section for more information.
Feel free to contact us – we are happy to help set up a sampling plan that will address the concerns you have.
A SNP Fingerprint can help you establish your new variety as separate from the parental lines, and it can be used to compare against in future cases.
We recommend testing a minimum of 3 separate plants of the new variety; or you can test 10 to give more confidence. You may also consider testing closely related varieties to confirm whether they can be differentiated using this test. If those varieties are already in our panel, then we can compare your new variety to our established fingerprints without the need to test them again.
Please note the limitations in testing sport mutants using SNP fingerprinting, which can be found in the FAQ’s.
SNP Fingerprinting works by comparing the fingerprint of your sample with other fingerprints of known plant varieties.
Qualterra maintains a reference database of fingerprints that we can compare your sample against; but if your variety of interest (your “reference”) is not included, you will need to test that variety in addition to your samples. If you wish, you may choose to add a reference even if it’s already listed in our database.
If you need to submit a reference, it is best practice to submit 3 samples from 3 different plants, but a single sample can also be used. Submitting 3 samples is important in cases where IP is being protected, but 1 sample is usually sufficient for projects confirming true-to-type.
Please contact us if you have questions about what kind of reference you need to submit. We are happy to walk through your project with you and get the best setup possible.
Apple Reference Varieties
| a3 268 7 | |
| Adams Apple | |
| Ambrosia | |
| Amere de Berthcourte | |
| Anna | |
| Antonovaka 1.5 | |
| Antonovaka Kamenichka | |
| Aurora | |
| Braeburn | |
| Bramtot | |
| Brookefield Gala | |
| Brown Snout | |
| Browns Apple | |
| Buckeye Gala | |
| Buckeye Prime | |
| Bud 9 | |
| Bulmer Norman | |
| Cap of Liberty | |
| Charden | |
| Chisel Jersey | |
| Cider Stayman | |
| Cider Stoke’s Red | |
| Cosmic Crisp | |
| Coxs Orange Pippin | |
| Crimson Beauty | |
| D16-78 10 | |
| Dabinette | |
| Delicious | |
| Domaine | |
| EarLleaf | |
| Early Cortland | |
| Emla 9 | |
| Esophus Spitzenberg | |
| Evercrisp® ‘MAIA-1’ | |
| Fuji Red Sport Type 2 | |
| Gala-USDA | |
| Geneva® 11 ‘G.11’ | |
| Geneva® 202 ‘G.202’ | |
| Geneva® 213 ‘G.213’ | |
| Geneva® 214 ‘G.214’ | |
| Geneva® 30 ‘G.30’ | |
| Geneva® CG.4004 | |
| Geneva® 41 ‘G.41’ | |
| Geneva® CG.5087 | |
| Geneva® CG.5257 | |
| Geneva® CG.6006 | |
| Geneva® 65 ‘G.65’ | |
| Geneva® 890 ‘G.890’ | |
| Geneva® 935 ‘G.935’ | |
| Geneva® 969 ‘G.969’ | |
| Golden Delicious | |
| Golden Harvey | |
| Golden Hornet | |
| Golden Russet | |
| Granny Smith | |
| Gravenstein | |
| Gravensteiner | |
| Grimes Golden | |
| Gros Frequin | |
| Harrison | |
| Harry Masters Jersey | |
| Hawkeye | |
| Honeycrisp | |
| Hudsons Golden Gem | |
| Indian Summer | |
| Jersey Mac | |
| Jonagold | |
| Jonathan | |
| Karma | |
| Kidds Orange Red | |
| King David | |
| Kingston Black | |
| Lady Williams | |
| Lodi | |
| M.11 | |
| M7 | |
| M9 | |
| M9-100 | |
| M9-337 | |
| M9-Troch | |
| Macoun | |
| Malus adirondack | |
| Mark | |
| McIntosh (Kimball) | |
| Melrose | |
| Mettais | |
| MM.111 | |
| MM.111-U | |
| Muscadet de Lense | |
| Mutsu | |
| Mutsu | |||
| Nic 29 | |||
| Oregon Spur Delicious | |||
| Otterson | |||
| Pajam-2 | |||
| Pazzaz | |||
| Pink Pearl | |||
| Prairifire | |||
| Priscilla | |||
| Red Delicious | |||
| Reine des Pommes | |||
| Rhode Island Greening | |||
| Roxbury Russet | |||
| Shizuka | |||
| Smokehouse | |||
| Spartan | |||
| Spigold | |||
| Spokane Beauty | |||
| Spy 227 | |||
| Stark’s Earliest | |||
| Sweet Alford | |||
| Sweet Coppin | |||
| Sweet Sixteen | |||
| Tomlan Sweet | |||
| Viking | |||
| Wickson | |||
| Worchester Pearmain | |||
| Yarlington Mill | |||
|
|||
Can other crops use this test?
No. SNP Fingerprinting only works on the crop it is listed for.
Can sports or bud- or limb-mutations be differentiated from the mother plant using SNP fingerprinting?
No. SNP Fingerprinting looks at only a few small pieces of the genome. Similarly, a sport mutant is usually a very small change in the genome – and the chances of the two being related is extremely low. If you would like to differentiate a sport mutant, contact us and we can direct you towards other testing methodologies that can be used to differentiate sports.
Can two similar varieties have the same fingerprint?
It’s unusual, but it is possible for two varieties to have the same fingerprint. There is about a 1/8000 chance of this happening. Fortunately, only a single difference between two fingerprints is needed to confirm that the two plants are not the same variety.
Can a SNP fingerprint tell us anything about its genetics or the traits of the plant?
No. SNP Markers that are used in fingerprinting are generally not associated with traits, to provide the maximum amount of discrimination power in the smallest number of tests. This means they are very useful for discriminating between varieties – but they don’t tell us anything about the traits of the plant.


